Table of contents
About
FerrDb is the world's first manually curated database for ferroptosis regulators and ferroptosis-disease associations from published journal articles.
This is the second version (FerrDb V2), a latest major update of the first version (FerrDb V1) published in 2020. Throughout the whole site, the terms FerrDb V2 and FerrDb are interchangeably used to indicate the second version. FerrDb will be regularly updated to support long-term service. The data in FerrDb is free to download and should be used for academic and research purposes only.
Ferroptosis is a new mode of regulated cell death that occurs as a consequence of iron-dependent accumulation of toxic lipid peroxidation. It has been implicated in a broad set of biological contexts, such as development, aging, and immunity. Also, it is tightly linked to a variety of human diseases, such as cancers, viral infections, and degenerative diseases. The ferroptotic process is complicated and consists of a wide range of metabolites and biomolecules. Even though great progress has been achieved, the mechanism of ferroptosis remains enigmatic. We are in an era of knowledge explosion, so it is of great importance to find ways to organize and utilize data efficiently. We noticed there lacks high-quality knowledgebase in ferroptosis research, so the database was created.
Data structure
| Data set | Primary category | Secondary category | Description |
|---|---|---|---|
| Driver | Regulator | Gene | A gene that promotes ferroptosis. |
| Suppressor | Regulator | Gene | A gene that prevents ferroptosis. |
| Marker | Regulator | Gene | A gene that indicates ferroptosis occurrence. |
| Unclassified gene | Regulator | Gene | A gene that is associated with ferroptosis but its regulatory role is unclear. It was assigned as Marker in FerrDb V1. |
| Inducer | Regulator | Substance | A chemical entity which enables to cause ferroptosis. |
| Inhibitor | Regulator | Substance | A chemical entity which enables to restrict ferroptosis. |
| Ferroptosis-disease association | Association | Disease association | Ferroptosis has two opposite effects on disease: aggravating an illness or alleviating an unpleasant situation. |
Confidence level
| Confidence | Explanation |
|---|---|
| Validated | The most reliable. It requires convincing evidence from strict tests such as pharmacological or genetic inhibition or activation test. |
| Screened | No strict validation. Only high-throughput data is available. |
| Predicted | No strict validation. In the source article, the authors drew their conclusions based on computational results or their knowledge. |
| Deduced | No strict validation. We, the curators, inferred the gene's function based on our understanding of the source article. |
Abbreviations
| Abbr. | Definition |
|---|---|
| _NA_ | Not available because of no resource data, or not applicable |
| System Xc- | Cystine/glutamate antiporter |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NOX | Nicotinamide adenine dinucleotide phosphate oxidase |
| GSH | Glutathione |
| alpha-KG | alpha-ketoglutarate |
| Gln | Glutamine |
| Glu | Glutamate |
| xCT | Light chain of System Xc- |
| PUFA(s) | Polyunsaturated fatty acid(s) |
| 5-HETE | 5-hydroxyeicosatetraenoic acid |
| PE | Phosphatidylethanolamine |
| AA | Arachidonoyl |
| AdA | Adrenoyl |
| LDH | Lactate dehydrogenase |
| 8-OHdG | 8-hydroxydeoxyguanosine |
| PUFAs | Polyunsaturated fatty acids |
| MDA | Lipid Peroxidation |
| MVB | Multivesicular body |
| RNS | Reactive nitrogen species |
Browse data
Click Browse in the navigation bar to open the drop-down menu that provides access to individual data set.
Data entries are shown on the page in a similar format. As shown in the following graph, each row is a regulator and attributes are shown in columns.
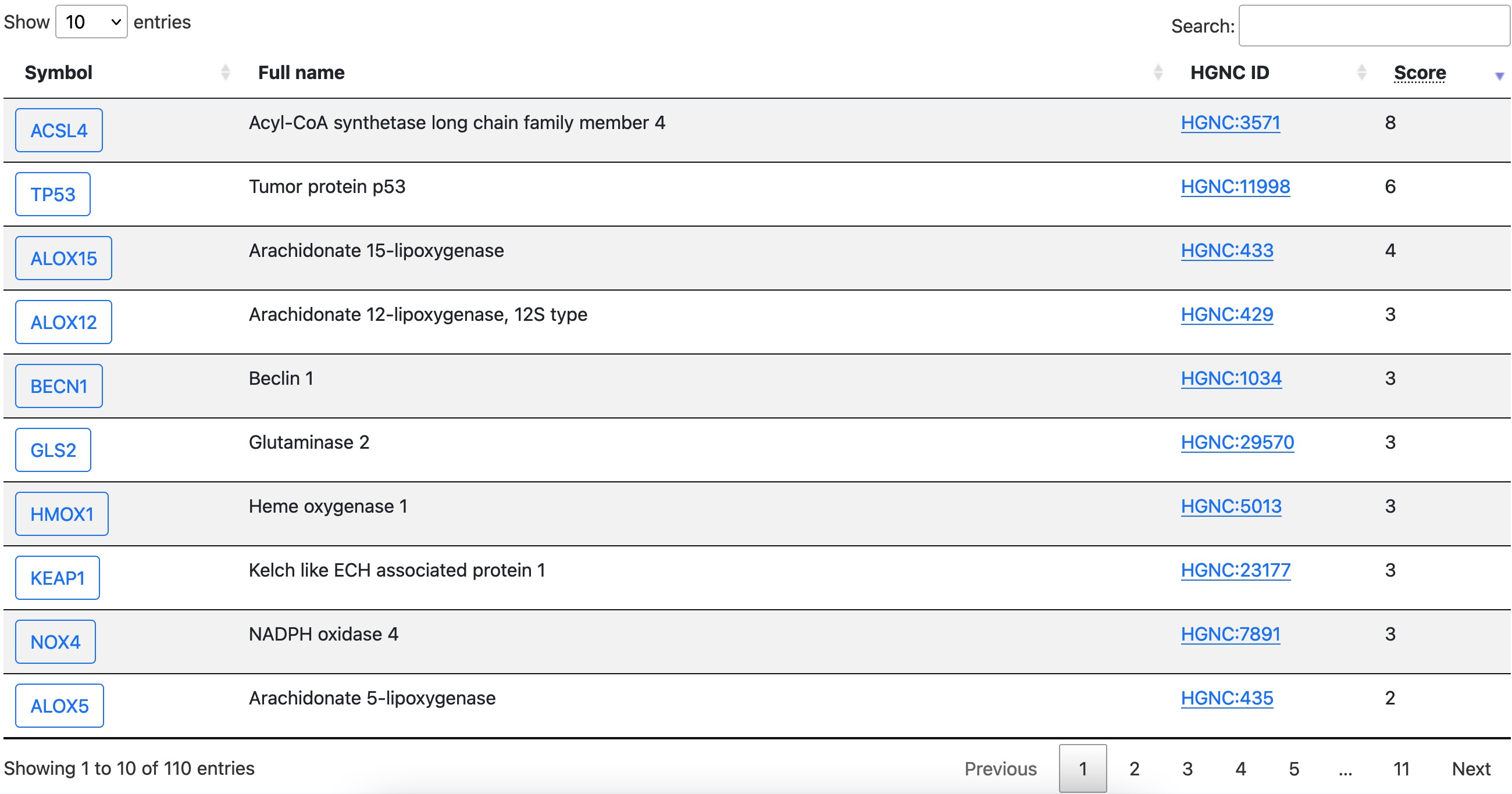
Click the symbol in the first column of this table to open the detail page of that gene.
The latest update column shows when the item was last updated. Please be notified that for items that is already in FerrDb V1, the latest update date is set to 2020/12/31. Since FerrDb V2, the latest update is set to the date of inclusion or modification of this item.
Download data
Please go to the Download page to get the data.
Submit data
We appreciate your contribution. Please go to the Submit page and follow the guidlines on that page to submit data to the database.
Use utilities
Regulation network
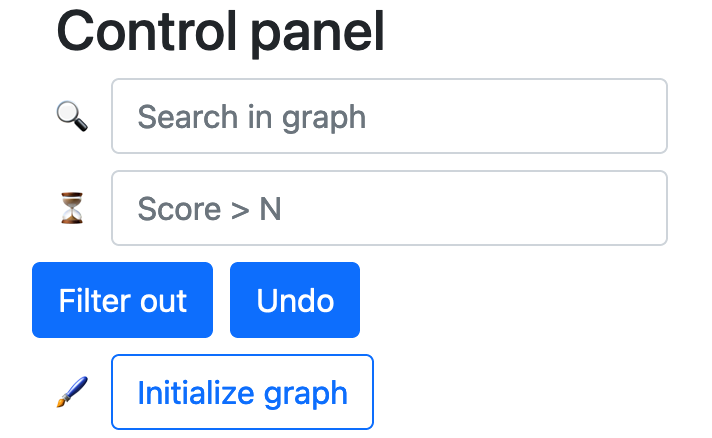
Under the Utilities drop-down, click Regulation network to open and use the utility. As shown in the example, black diamond denotes ferroptosis regulator (e.g., gene), grey circle denotes other involved regulatory elements, orange line with a tee head represents inhibiting effect, and grass green line with an arrow head represents promoting effect. The size of diamond indicates experiment counts. The more the number of experiments, the larger the node size.

Users can play with the regulation network with tools from the control panel, such as finding a gene of their interest and filtering out genes with score > N. If you unfortunately mess up things, no worries, you can press the Initialize graph button to get the page tidy.
Enrichment analysis
Enrichment analysis enables users to know whether a gene list of their interest is statistically enriched in any ferroptosis gene set annotated in FerrDb. The analysis is performed using GSEApy, with both Enrichr's over-representation and Subramanian et al.'s GSEA methods available.
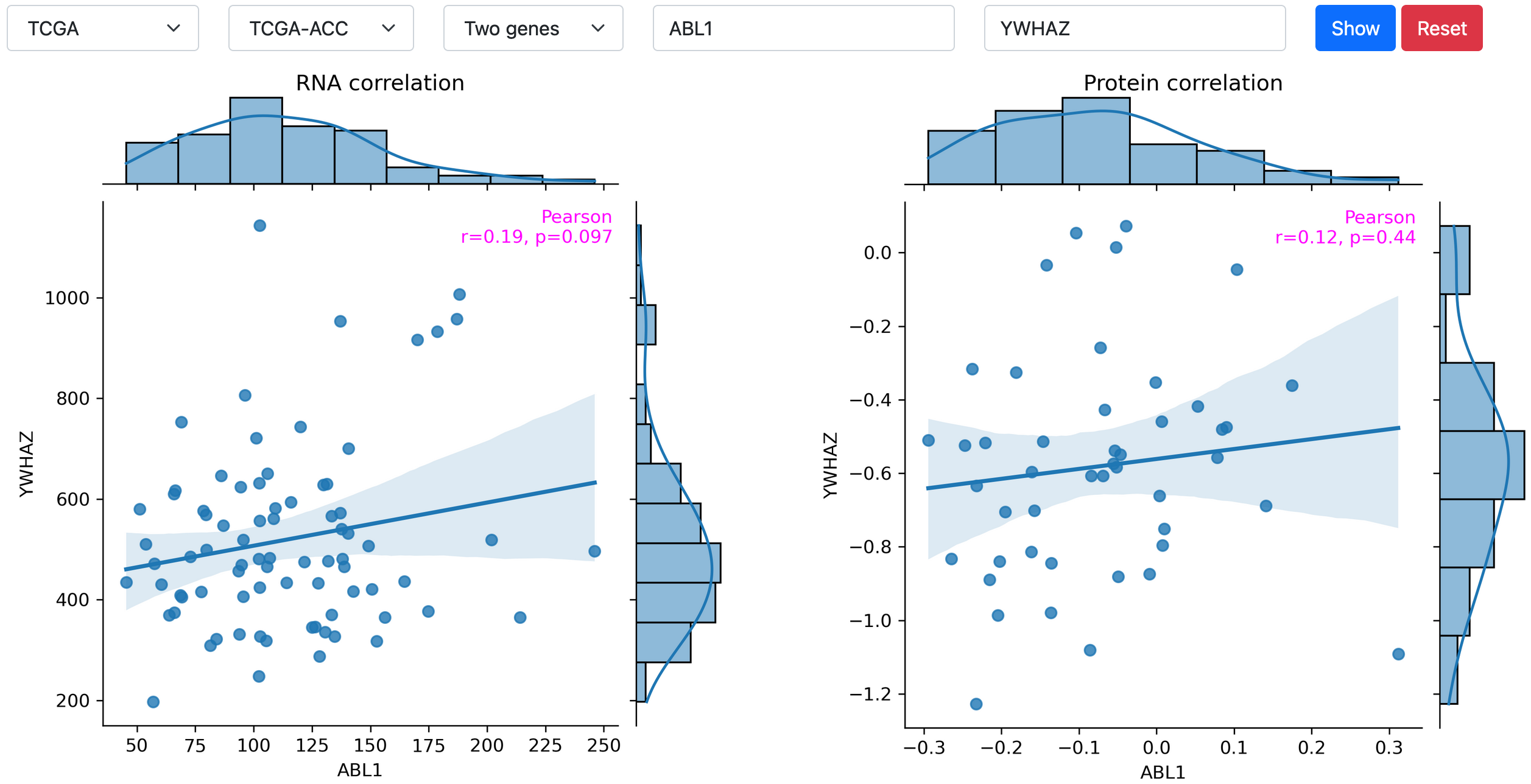
Correlation analysis
This utility can be used for two purposes: (i) to see the correlation between RNA levels and protein levels of a gene; (ii) to see the correlation between two genes on the RNA or protein level. As shown in the following example, the Pearson's r and p value are denoted.
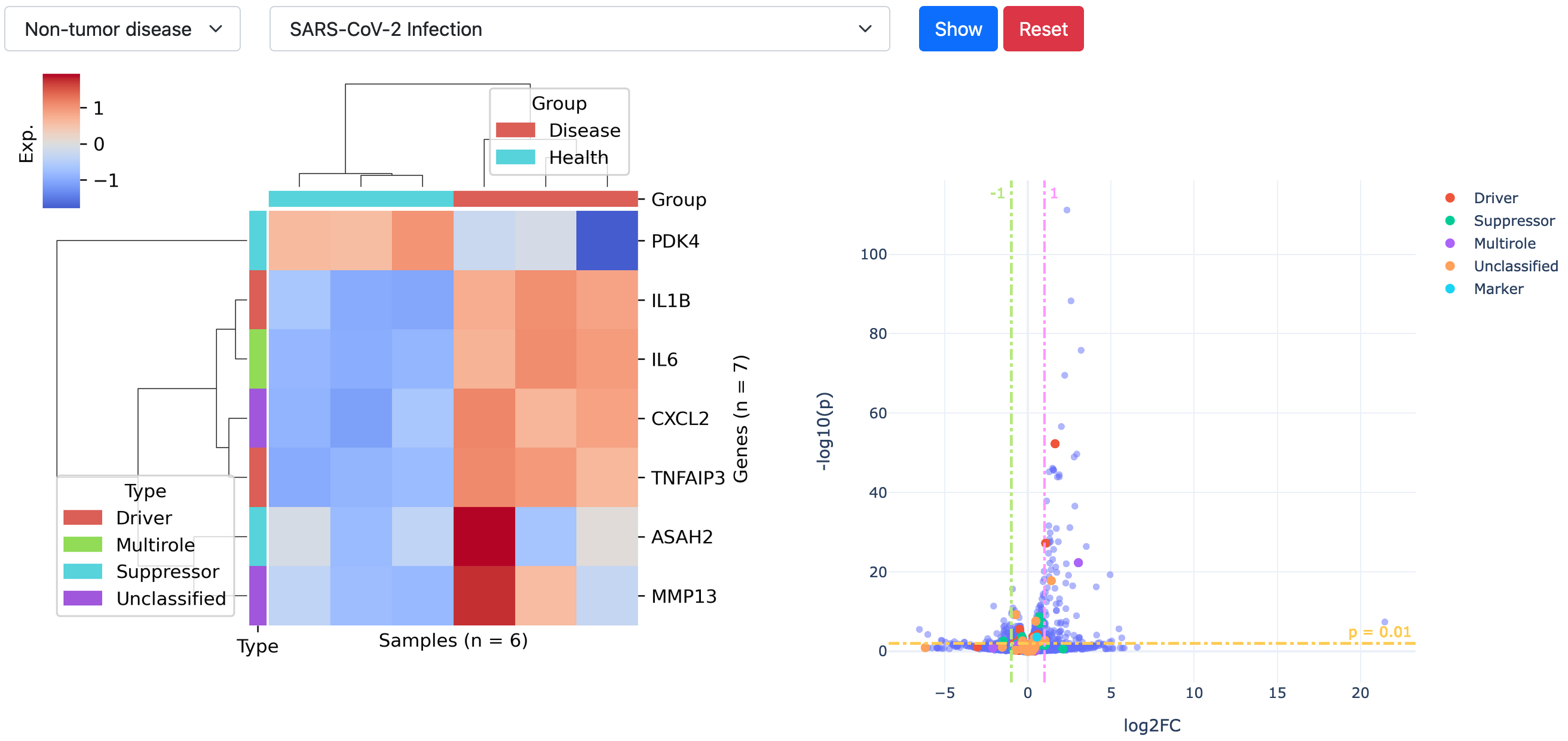
DEG landscape
This utility can be used to see differential expression patterns of all ferroptosis gene regulators in a disease at once. As shown in the example below, the DEG landscape can be created after setting disease type and name.
Multirole regulator
Use this utility to browse genes that regulate ferroptosis in multiple ways.
Feedback
We wish to hear from you. Please send an email to the webmaster at admin@zhounan.org, if you have any comments.
Working group
| Name | Task/Role | Affiliation |
|---|---|---|
| Nan Zhou | Article collection | The Affiliated Brain Hospital of Guangzhou Medical University |
| Curation and annotation | ||
| Coding | ||
| Coordinator | ||
| Yuping Ning | Quality control | The Affiliated Brain Hospital of Guangzhou Medical University |
| Qingsong Du | Curation and annotation | Sichuan University |
| Zhiyu Zhang | Curation and annotation | Sichuan University |
| Jinku Bao | Quality control | Sichuan University |
| Xiaoqing Yuan | Curation and annotation | Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University |
| Li Peng | Data management | Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University |