FerrDb is the world’s first manually curated database for regulators and markers of ferroptosis and ferroptosis-disease associations. It will be routinely updated to support long-term service. The data in FerrDb is free to download and use.
Ferroptosis is a new mode of regulated cell death that depends on iron. Cells die from the toxic accumulation of lipid reactive oxygen species inside. It is tightly linked to a variety of human diseases, such as cancers and degenerative diseases. The ferroptotic process is complicated and consists of a wide range of metabolites and biomolecules. Even though great progress has been achieved, the mechanism of ferroptosis remains enigmatic. We are in an era of knowledge explosion, so it is of importance to find ways to organize and utilize data efficiently. We noticed there lacks high-quality knowledgebase in ferroptosis research, so the database was created.
| Abbr. | Definition |
|---|---|
| _NA_ | Not available because of no resource data, or not applicable |
| System Xc- | Cystine/glutamate antiporter |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NOX | Nicotinamide adenine dinucleotide phosphate oxidase |
| GSH | Glutathione |
| alpha-KG | alpha-ketoglutarate |
| Gln | Glutamine |
| Glu | Glutamate |
| xCT | Light chain of System Xc- |
| PUFA(s) | Polyunsaturated fatty acid(s) |
| 5-HETE | 5-hydroxyeicosatetraenoic acid |
| PE | Phosphatidylethanolamine |
| AA | Arachidonoyl |
| AdA | Adrenoyl |
| LDH | Lactate dehydrogenase |
| 8-OHdG | 8-hydroxydeoxyguanosine |
| PUFAs | Polyunsaturated fatty acids |
| MDA | Lipid Peroxidation |
| MVB | Multivesicular body |
| RNS | Reactive nitrogen species |
| Confidence | Explanation |
|---|---|
| Validated | The most reliable. It requires convincing evidence from strict tests such as pharmacological or genetic inhibition or activation test. |
| Screened | No strict validation. Only high-throughput data is available. |
| Predicted | No strict validation. In the source article, the original authors drew their conclusions based on computational results or their knowledge. |
| Deduced | No strict validation. We, the curators, inferred the gene's function based on our understanding of the original article. |
| Term | Definition |
|---|---|
| Regulator | A gene or small molecule that participants in the regulation of ferroptosis. |
| Driver | A gene that promotes ferroptosis. |
| Suppressor | A gene that prevents ferroptosis. |
| Marker | A gene that indicates the occurrence of ferroptosis. |
| Inducer | A small molecule which enables to cause ferroptosis. |
| Inhibitor | A small molecule which enables to restrict ferroptosis. |
Type search terms in the search box and press enter to search FerrDb. By default, FerrDb matches gene. If you want to match any text, please select the "All" radio button.
Hover your mouse pointer on "Browse" and you will see a drop-down menu which provides access to different annotation data sets.
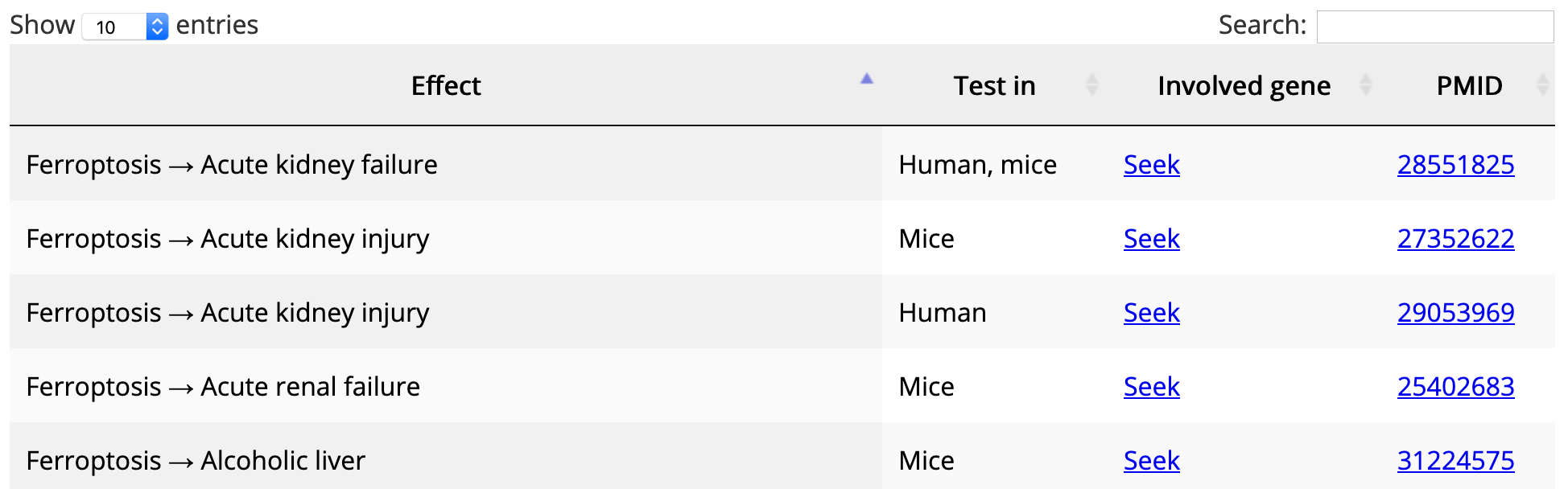
In the data table, items are listed by row. Table headers can be clicked to sort rows. Table content is searchable via the search box on the top right of the table.
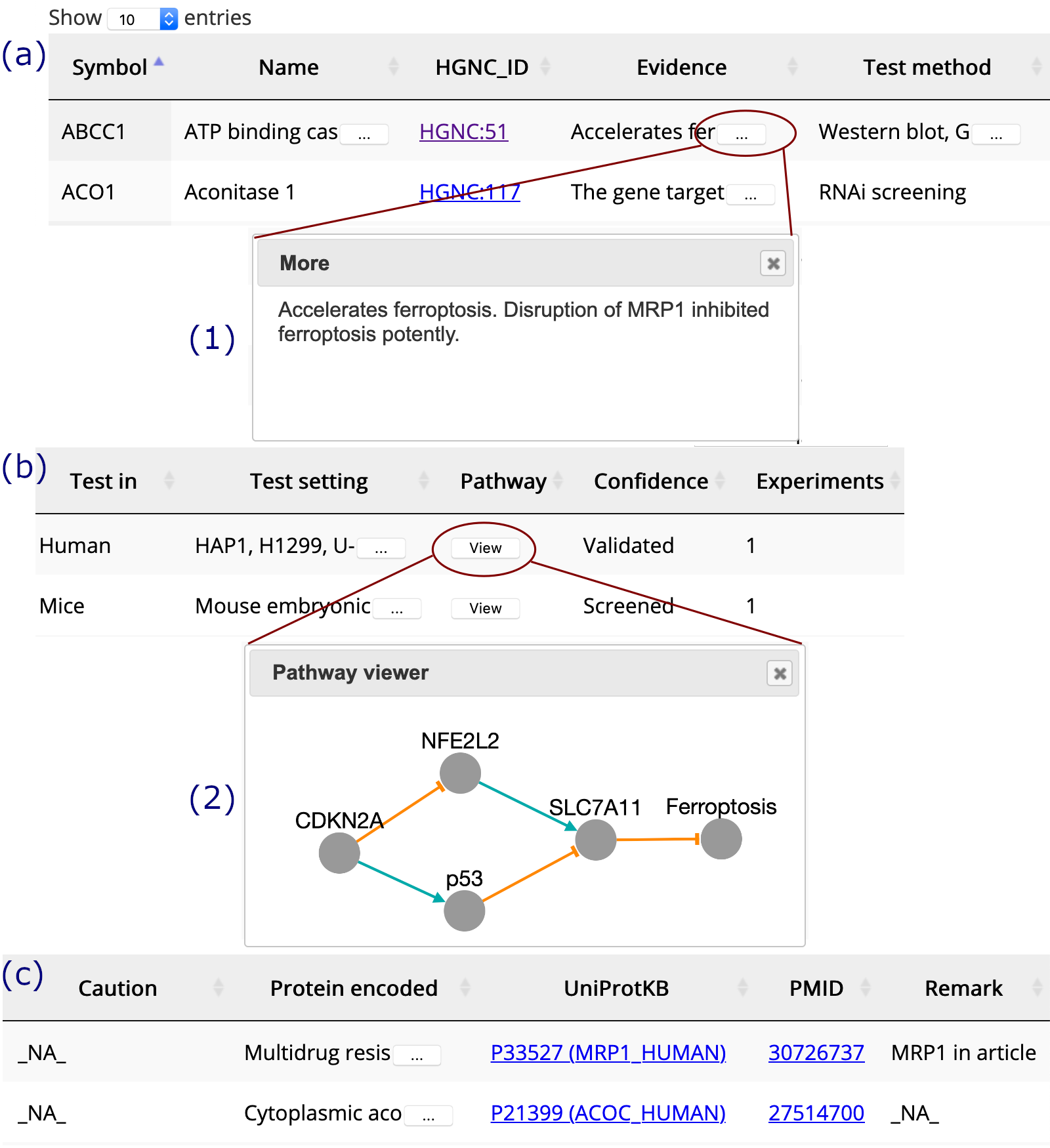
If you see text truncated, please click on the "ellipsis" button to read the full content. The "view" button in the Pathway column is provided to visualize gene's regulation pathway.
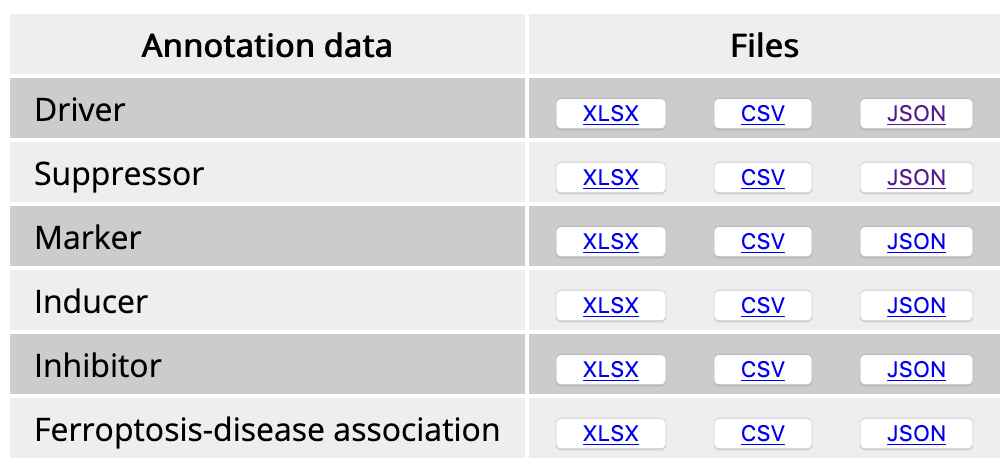
Click on "Download" in the navigation bar and then downloadable files are shown on the download page.
Hover the mouse pointer on "Utilities" and click "Regulation network".
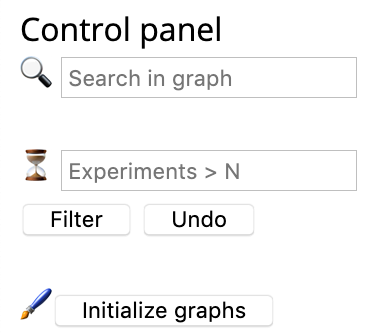
Then a regulation network will show. Black diamond denotes ferroptosis regulator (e.g., gene), grey circle denotes other regulatory elements involved, orange line with a tee head represents promoting effect, and grass green line with an arrow head represents inhibiting effect. The size of diamond indicates experiment count. The more the number of experiments, the larger the node size.

You can play with the regulation network with tools from the control panel, such as finding a gene of your interest and filtering out genes with more than N experiments. If you unfortunately mess up things, no worries, you can press the "Initialize graphs" button to get your page tidy.
To check the consistency of gene symbols in FerrDb with HGNC, hover the mouse pointer on "Utilities" and click "Gene symbol validation".
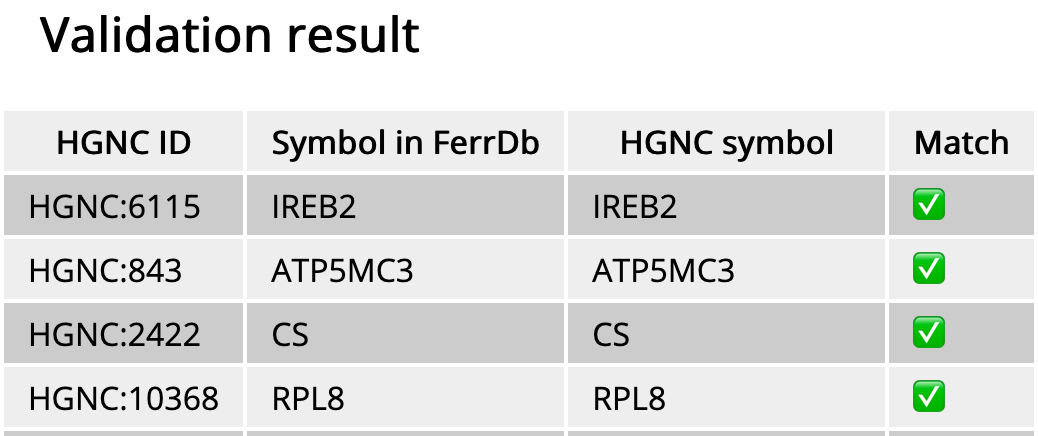
Then, gene symbols will be checked against HGNC using the genenames.org REST API service. If a gene symbol is not the same in FerrDb and HGNC, it is marked with ❌, otherwise it is marked with ✅. When you find a discrepancy remains unfixed for two weeks, please notify us and then we will fix it in our database.
The small molecule images contained in FerrDb are from result pages of Microsoft Bing search engine. Credits are given to the creators. If any of the images belong to you and you want us to remove them, please let us know (admin@zhounan.org). FerrDb does not own or claims to own these images, whoever wants to reuse these images needs to ask for permission from the owners or copyright holders.
We value your voice. If you have any suggestions for us or if you want to join this program to enrich this database, please contact the maintainer via admin@zhounan.org.