Contents
- Motivation of IntApop
- What can IntApop do?
- Link/interaction levels in IntApop
- Model organisms in IntApop
- Data in IntApop
- Prediction cutoff
- Prediction samples
- Query samples
- Large data prediction
- License
Motivation of IntApop
Apoptosis, or type I programmed cell death, plyas pivotal role in controlling cell number and proliferation as a part of normal development through a complex network of molecules that mediate death and survival signals. Protein-protein interactions (PPIs) and signal transduction pathways are essential for apoptosis processes. Proteins interact with each other specifically and selectively to achieve their biological functions. Consequently, establishing apoptotic protein interaction networks is crucial for systematically understanding apoptosis, especially its signaling pathways. However, the convenient and necessary web servers for apoptotic protein interactions prediction are still unavailable now. So we make an initial step to develop the IntApop for apoptotic protein interactions prediction and visualization in human. To cite IntApop, please reference: N. Zhou, J. Zhang, L. Feng, etc., Biosystems 2013 DOI: 10.1016/j.biosystems.2013.09.007. The original publication is N. Zhou, J. Zhang, L. Feng, etc., IntApop: A web service for predicting apoptotic protein interactions in humans. Biosystems 2013;114(3):238-244.
What can IntApop do?
IntApoop can be used to predict potential apoptotic protein interactions, visualize predicted interactions and, query global human apoptotic interactions network.
Link/interaction levels in IntApop
IntApop provides two levels of link analyses for the query protein pairs:
- known association: which comes from other protein interactions databases.
- predicted association: which is delivered by IntApop.
Model organisms in IntApop
Homo sapiens (human)
The only living species in the Homo genus of bipedal primates in Hominidae, the great ape family.
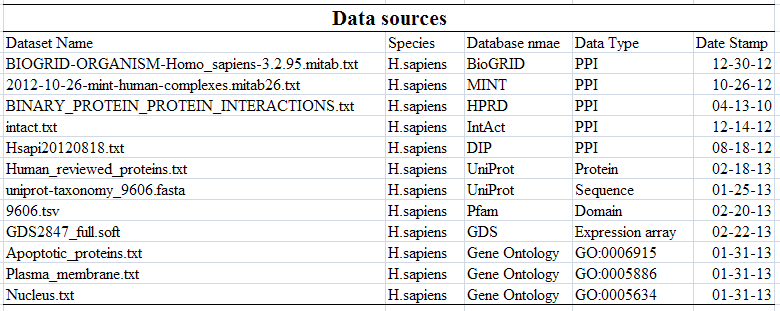
- Existed human PPIs from DIP, IntAct, HPRD, BioGRID and MINT.
- Reviewed human proteins come from UniProtKB.
- Apoptosis proteins are from GO: 0006915.
Prediction cutoff
Prediction cutoff is very important in interactions prediction.
IntApop uses it to determine whether two proteins interact or not.
You can select a cutoff according to the picture below:
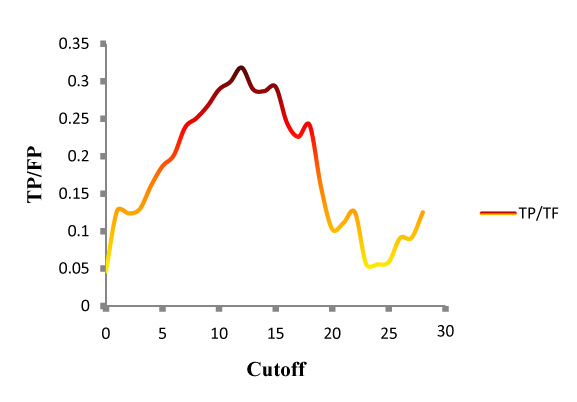
The picture above shows the TP/FP ratios at defferent cutoffs, indicating that the maximum TP/FP is at 12.
Indeed, we use 12 as the optimal threshold in our study and it is selected as the default cutoff for IntApop prediction.
Prediction samples
1. Refer to the prediction page.
2. Choose a method: input prediction or upload prediction.
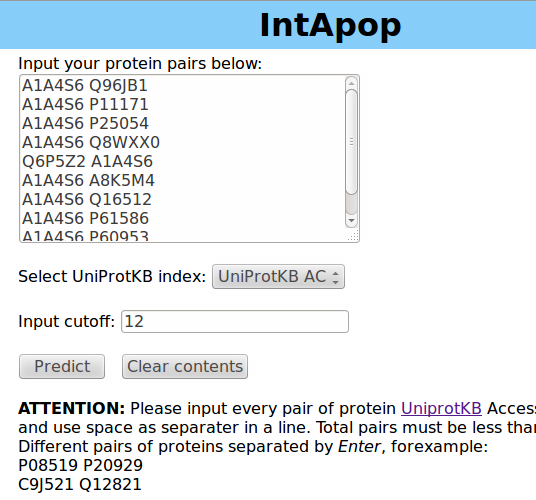
2.1 If the number of your protein pairs is less than 100, please use "input prediction", for example:
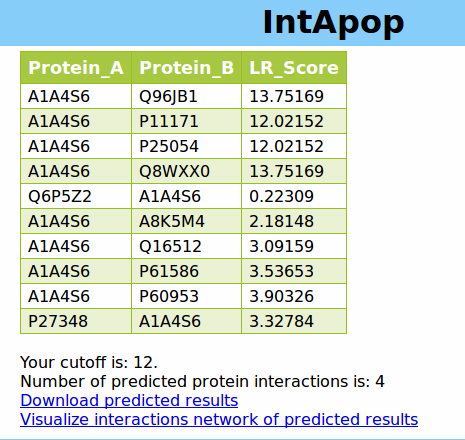
2.1.1 Predict results:
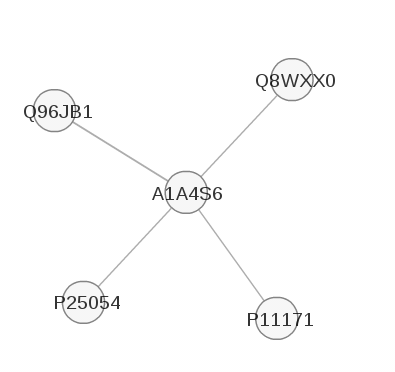
2.1.2 Visualizing predicted interactions:
2.2 If you have more than 100 items, please upload a properly formatted file to predict.
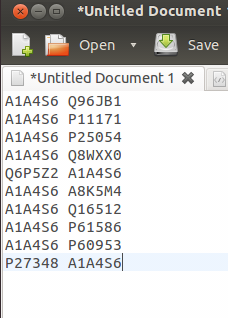
2.2.1 File format example:
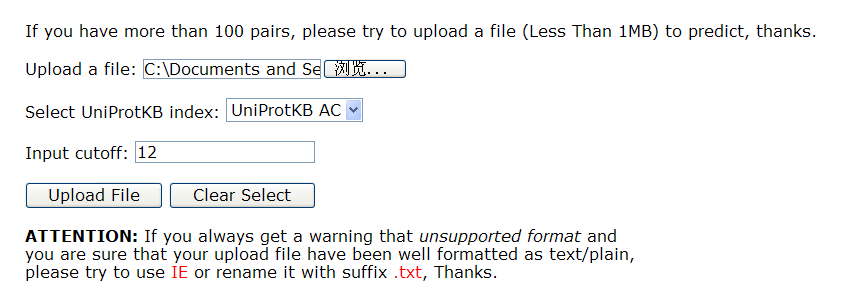
2.2.2 Upload your file and predict:
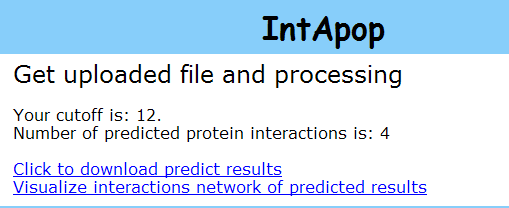
2.2.3 Predict results:
Query samples
Users can input UniProtKB AC or ID to query and visualize interactions. For example O00755:
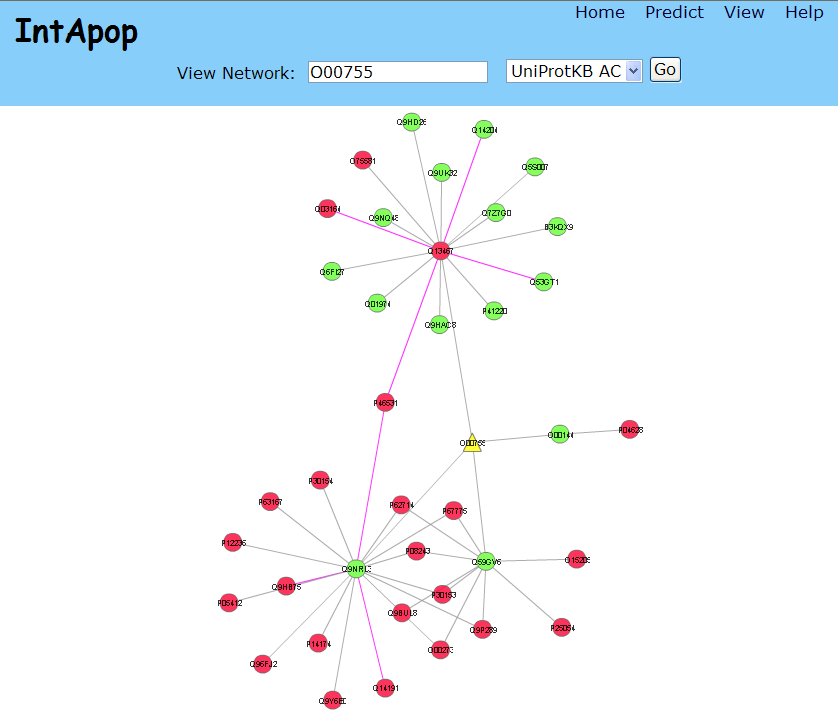
In picture above, nodes represent proteins and edges represent associations and:
- Yellow triangle(s) is(are) queried protein(s).
- Red nodes are apoptotic proteins.
- Green nodes are other proteins.
- Gray edges are known associations.
- Pink edges are predicted associations.
Large data prediction
In order to guarantee the performance of IntApop, the upload file size is limited to 1MB. Otherwise, you can send an email to IntApop omnibus for large data prediction.
License
Of note, IntApop is freely avaliable to any user wishing to use it for non-commercial purpose, otherwise you must contact us to get the permission.